library(marginaleffects)
library(patchwork)
library(lme4)
dat <- mtcars
dat$cyl <- factor(dat$cyl)
dat$am <- as.logical(dat$am)
mod <- lmer(mpg ~ hp + am + (1 | cyl), data = dat)34 Uncertainty
This vignette is a complement to the core chapter on hypothesis testing: Chapter 14 It presents more advanced examples.
34.1 Mixed effects models: Satterthwaite and Kenward-Roger corrections
For linear mixed effects models we can apply the Satterthwaite and Kenward-Roger corrections in the same way as in previous chapters:
Marginal effects at the mean with classical standard errors and z-statistic:
slopes(mod, newdata = "mean")
#>
#> Term Contrast Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> am TRUE - FALSE 4.6661 1.1343 4.11 <0.001 14.6 2.4430 6.8892
#> hp dY/dX -0.0518 0.0115 -4.52 <0.001 17.3 -0.0743 -0.0294
#>
#> Type: responseMarginal effects at the mean with Kenward-Roger adjusted variance-covariance and degrees of freedom:
slopes(mod,
newdata = "mean",
vcov = "kenward-roger")
#>
#> Term Contrast Estimate Std. Error t Pr(>|t|) S 2.5 % 97.5 % Df
#> am TRUE - FALSE 4.6661 1.2824 3.64 0.0874 3.5 -1.979 11.3113 1.68
#> hp dY/dX -0.0518 0.0152 -3.41 0.0965 3.4 -0.131 0.0269 1.68
#>
#> Type: responseWe can use the same option in any of the package’s core functions, including:
plot_predictions(mod, condition = "hp", vcov = "satterthwaite")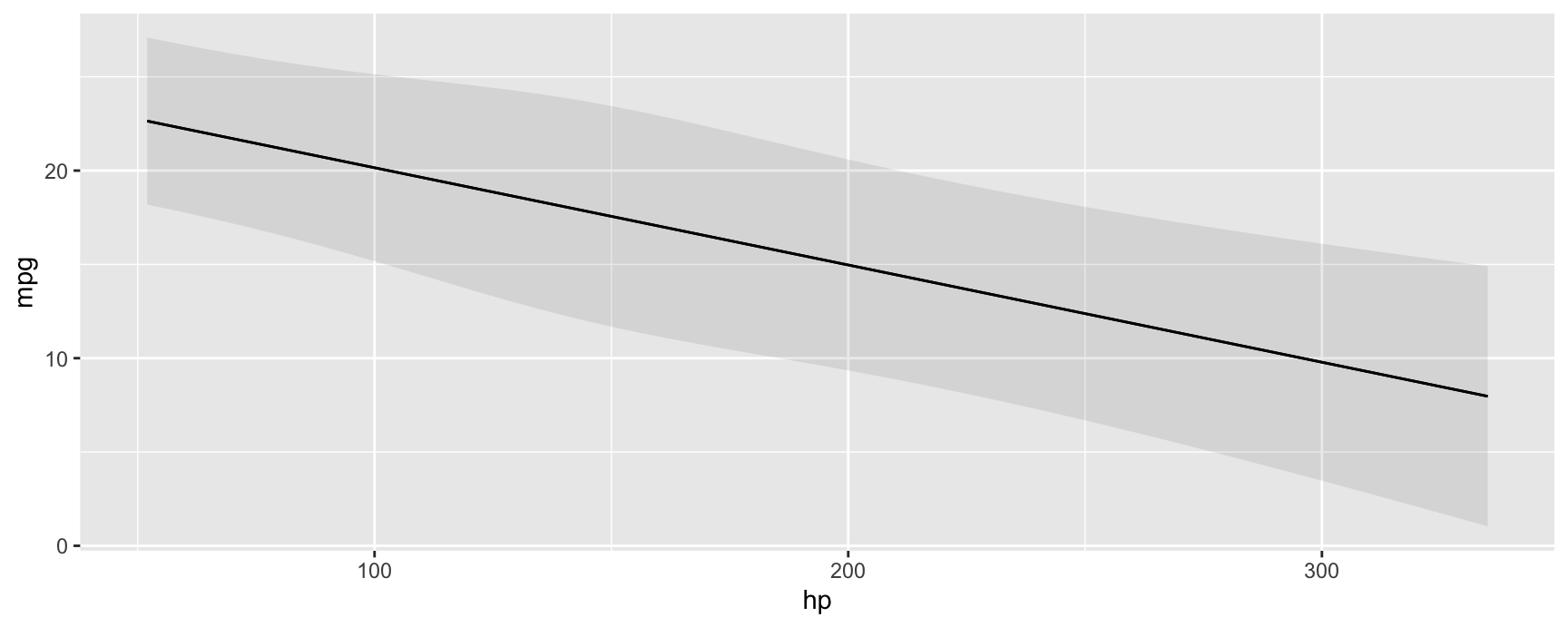
34.2 Numerical derivatives: Sensitivity to step size
dat <- get_dataset("penguins", "palmerpenguins")
dat$large_penguin <- ifelse(dat$body_mass_g > median(dat$body_mass_g, na.rm = TRUE), 1, 0)
mod <- glm(large_penguin ~ bill_length_mm * flipper_length_mm + species, data = dat, family = binomial)marginaleffects uses numerical derivatives in two contexts:
- Estimate the partial derivatives reported by
slopes()function.- Centered finite difference
-
\(\frac{f(x + \varepsilon_1 / 2) - f(x - \varepsilon_1 / 2)}{\varepsilon_1}\), where we take the derivative with respect to a predictor of interest, and \(f\) is the
predict()function.
- Estimate standard errors using the delta method.
- Forward finite difference
-
\(\frac{g(\hat{\beta}) - g(\hat{\beta} + \varepsilon_2)}{\varepsilon_2}\), where we take the derivative with respect to a model’s coefficients, and \(g\) is a
marginaleffectsfunction which returns some quantity of interest (slopes, contrasts, predictions, etc.)
Note that the step sizes used in those two contexts can differ. If the variables and coefficients have very different scales, it may make sense to use different values for \(\varepsilon_1\) and \(\varepsilon_2\).
By default, \(\varepsilon_1\) is set to 1e-4 times the range of the variable with respect to which we are taking the derivative. By default, \(\varepsilon_2\) is set to the maximum value of 1e-8, or 1e-4 times the smallest absolute coefficient estimate. (These choices are arbitrary, but I have found that in practice, smaller values can produce unstable results.)
\(\varepsilon_1\) can be controlled by the eps argument of the slopes() function. \(\varepsilon_2\) can be controlled by setting a global option which tells marginaleffects to compute the jacobian using the numDeriv package instead of its own internal functions. This allows more control over the step size, and also gives access to other differentiation methods, such as Richardson’s. To use numDeriv, we define a list of arguments which will be pushed forward to numDeriv::jacobian:
avg_slopes(mod, variables = "bill_length_mm")
#>
#> Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0.0279 0.00595 4.69 <0.001 18.4 0.0162 0.0395
#>
#> Term: bill_length_mm
#> Type: response
#> Comparison: dY/dX
options(marginaleffects_numDeriv = list(method = "Richardson"))
avg_slopes(mod, variables = "bill_length_mm")
#>
#> Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0.0279 0.00595 4.69 <0.001 18.4 0.0162 0.0395
#>
#> Term: bill_length_mm
#> Type: response
#> Comparison: dY/dX
options(marginaleffects_numDeriv = list(method = "simple", method.args = list(eps = 1e-3)))
avg_slopes(mod, variables = "bill_length_mm")
#>
#> Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0.0279 0.568 0.049 0.961 0.1 -1.09 1.14
#>
#> Term: bill_length_mm
#> Type: response
#> Comparison: dY/dX
options(marginaleffects_numDeriv = list(method = "simple", method.args = list(eps = 1e-5)))
avg_slopes(mod, variables = "bill_length_mm")
#>
#> Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0.0279 0.00601 4.64 <0.001 18.1 0.0161 0.0396
#>
#> Term: bill_length_mm
#> Type: response
#> Comparison: dY/dX
options(marginaleffects_numDeriv = list(method = "simple", method.args = list(eps = 1e-7)))
avg_slopes(mod, variables = "bill_length_mm")
#>
#> Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
#> 0.0279 0.00595 4.68 <0.001 18.4 0.0162 0.0395
#>
#> Term: bill_length_mm
#> Type: response
#> Comparison: dY/dXNotice that the standard errors can vary considerably when using different step sizes. It is good practice for analysts to consider the sensitivity of their results to this setting.
Now, we illustrate the full process of standard error computation, using raw R code. First, we choose two step sizes:
eps1 <- 1e-5 # slope
eps2 <- 1e-7 # delta method
s <- slopes(mod, newdata = head(dat, 3), variables = "bill_length_mm", eps = eps1)
print(s[, 1:5], digits = 6)
#>
#> Estimate Std. Error
#> 0.0179765 0.00881402
#> 0.0359630 0.01254629
#> 0.0849071 0.02137500
#>
#> Term: bill_length_mm
#> Comparison: dY/dXWe can get the same estimates manually with these steps:
linkinv <- mod$family$linkinv
## increment the variable of interest by h
dat_hi <- transform(dat, bill_length_mm = bill_length_mm + eps1)
## model matrices: first 3 rows
mm_lo <- insight::get_modelmatrix(mod, data = dat)[1:3,]
mm_hi <- insight::get_modelmatrix(mod, data = dat_hi)[1:3,]
## predictions
p_lo <- linkinv(mm_lo %*% coef(mod))
p_hi <- linkinv(mm_hi %*% coef(mod))
## slopes
(p_hi - p_lo) / eps1
#> [,1]
#> 1 0.01797653
#> 2 0.03596304
#> 3 0.08490712To get standard errors, we build a jacobian matrix where each column holds derivatives of the vector valued slope function, with respect to each of the coefficients. Using the same example:
This gives us the first column of \(J\), which we can recover in full from the marginaleffects object attribute:
J <- components(s, "jacobian")
J
#> (Intercept) bill_length_mm flipper_length_mm speciesChinstrap speciesGentoo bill_length_mm:flipper_length_mm
#> [1,] 0.01601497 0.6777495 2.897231 0 0 122.6914
#> [2,] 0.02770006 1.1957657 5.153100 0 0 222.4989
#> [3,] 0.02281508 1.1500800 4.440004 0 0 224.0830To build the full matrix, we would simply iterate through the coefficients, incrementing them one after the other. Finally, we get standard errors via:
Which corresponds to our original standard errors:
print(s[, 1:5], digits = 7)
#>
#> Estimate Std. Error
#> 0.01797650 0.008814015
#> 0.03596299 0.012546293
#> 0.08490708 0.021375005
#>
#> Term: bill_length_mm
#> Comparison: dY/dXReverting to default settings:
options(marginaleffects_numDeriv = NULL)Note that our default results for this model are very similar – but not exactly identical – to those generated by the margins. As should be expected, the results in margins are also very sensitive to the value of eps for this model:
library(margins)
margins(mod, variables = "bill_length_mm", data = head(dat, 3), unit_ses = TRUE)$SE_dydx_bill_length_mm
margins(mod, variables = "bill_length_mm", data = head(dat, 3), eps = 1e-4, unit_ses = TRUE)$SE_dydx_bill_length_mm
margins(mod, variables = "bill_length_mm", data = head(dat, 3), eps = 1e-5, unit_ses = TRUE)$SE_dydx_bill_length_mm34.3 Bayesian estimates and credible intervals
See the brms vignette for a discussion of bayesian estimates and credible intervals.