library(marginaleffects)
library(tinyplot)
tinytheme("ipsum")
set.seed(48103)
# number of days
n <- 365
# intervention at day
intervention <- 200
# time index from 1 to 365
time <- c(1:n)
# treatment variable: 0 before the intervention and 1 after
treatment <- c(rep(0, intervention), rep(1, n - intervention))
# outcome equation
outcome <-
10 + # pre-intervention intercept
15 * time + # pre-intervention slope (trend)
20 * treatment + # post-intervention intercept (shift of 10)
5 * treatment * time + # steeper slope after the intervention
rnorm(n, mean = 0, sd = 100) # noise
dat <- data.frame(outcome, time, treatment)24 Interrupted Times Series
Interrupted Time Series (ITS) analysis is a widely used research design that aims to infer the impact of a discrete intervention or event by examining changes in an outcome measure over time. It finds applications in various fields, including public health policies—such as evaluating the effect of vaccination campaigns—and educational settings—like assessing curriculum reform. The key motivation behind ITS is to leverage data already being collected, identify a clear “interruption” at a known point in time (e.g., when a policy was enacted), and then compare the behavior of the outcome measure before and after this interruption.
Although ITS can be extremely useful, it also poses substantial methodological and practical challenges. Unlike experimental or quasi-experimental approaches, ITS relies on very strong assumptions about the continuity or predictability of the outcome’s pre-intervention trajectory. Unobserved confounding events and gradual policy rollouts can violate these assumptions, complicating efforts to attribute changes solely to the interruption.
ITS shares some similarities with the popular difference-in-differences (DiD) approach, in which outcomes in a treated group are contrasted with those in an untreated comparison group. Meeting the assumptions required for credible inference with DiD is extremely challenging, as is attested by recent methodological developments in this field (Roth et al. 2023). ITS is arguably even more difficult to get right, since it typically relies only on a single times series, and thus lacks a true control group.
This article shows how to use the marginaleffects package to analyze a simple ITS setup. To illustrate, we consider a dataset simulated by this code:
The data look like this:
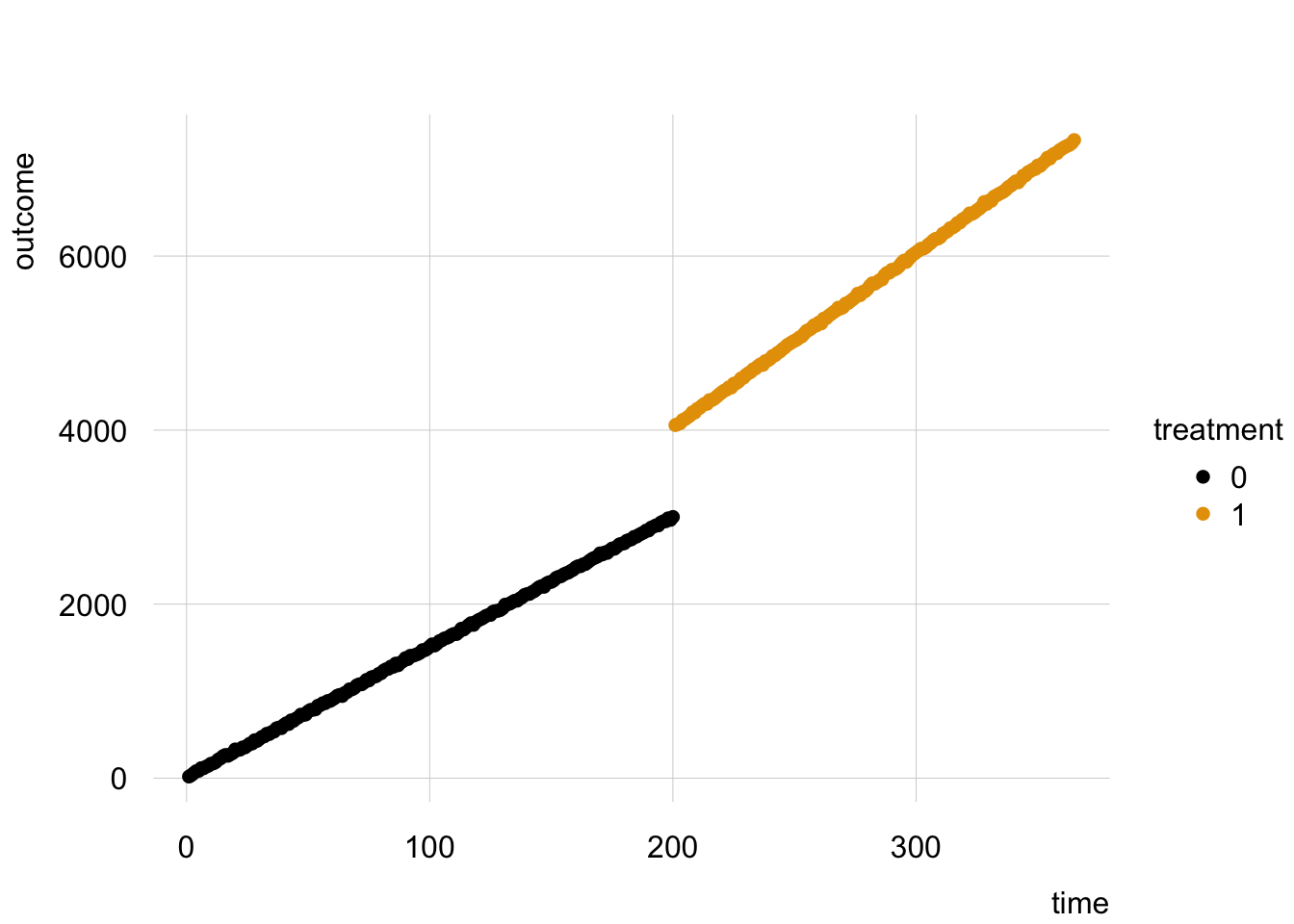
Often, analysts will model such data using a linear regression model with multiplicative interaction, and a specially constructured “time since intervention” variable:
\[ \text{Outcome} = \beta_0 + \beta_1 \text{Time} + \beta_2 \text{Treatment} + \beta_3 \text{Time since intervention} + \varepsilon \]
With marginaleffects, we do not even need to construct this special variable.1 All we have to do is fit this mathematically equivalent—and simpler—model:
\[ \text{Outcome} = \alpha_0 + \alpha_1 \text{Time} + \alpha_2 \text{Treatment} + \alpha_3 \text{Time} \times \text{Treatment} + \varepsilon \]
Call:
lm(formula = outcome ~ time * treatment, data = dat)
Residuals:
Min 1Q Median 3Q Max
-261.551 -69.865 0.908 66.543 308.257
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.4518 13.6298 0.327 0.744
time 15.0023 0.1176 127.574 <2e-16 ***
treatment -17.6430 47.0542 -0.375 0.708
time:treatment 5.1581 0.1961 26.303 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 96.02 on 361 degrees of freedom
Multiple R-squared: 0.9982, Adjusted R-squared: 0.9982
F-statistic: 6.804e+04 on 3 and 361 DF, p-value: < 2.2e-16Then, we can use the marginaleffects package to answer a series of scientific questions. For example, what would be the predicted outcome at time 200 with or without intervention?
p0 <- predictions(mod, newdata = datagrid(time = 199, treatment = 0))
p1 <- predictions(mod, newdata = datagrid(time = 200, treatment = 1))
p0
time treatment Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
199 0 2990 13.4 223 <0.001 Inf 2964 3016
Type: responsep1
time treatment Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
200 1 4019 15 268 <0.001 Inf 3989 4048
Type: responseThis suggests that the intervention would have increased the outcome by about 1028.9766399 units at time 200. Is the difference between those predictions statistically significant?
comparisons(mod, variables = "treatment", newdata = datagrid(time = 200))
time Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
200 1014 20.2 50.2 <0.001 Inf 974 1054
Term: treatment
Type: response
Comparison: 1 - 0What is the average slope of the outcome equation with respect to time, before and after the intervention?
avg_slopes(mod, variables = "time", by = "treatment")
treatment Estimate Std. Error z Pr(>|z|) S 2.5 % 97.5 %
0 15.0 0.118 128 <0.001 Inf 14.8 15.2
1 20.2 0.157 128 <0.001 Inf 19.9 20.5
Term: time
Type: response
Comparison: dY/dXThis shows that the outcome increases more rapidly after the intervention.
Many analysts like to visualize the results of an ITS analysis, by plotting the predicted outcome for the control group, knowing that these predictions are counterfactual after the intervention. This can be done by first make predictions for all combinations of time and treatment, dropping the observations we do not want to plot, and feeding the data to a plotting package like ggplot2.
Warning: package 'ggplot2' was built under R version 4.5.2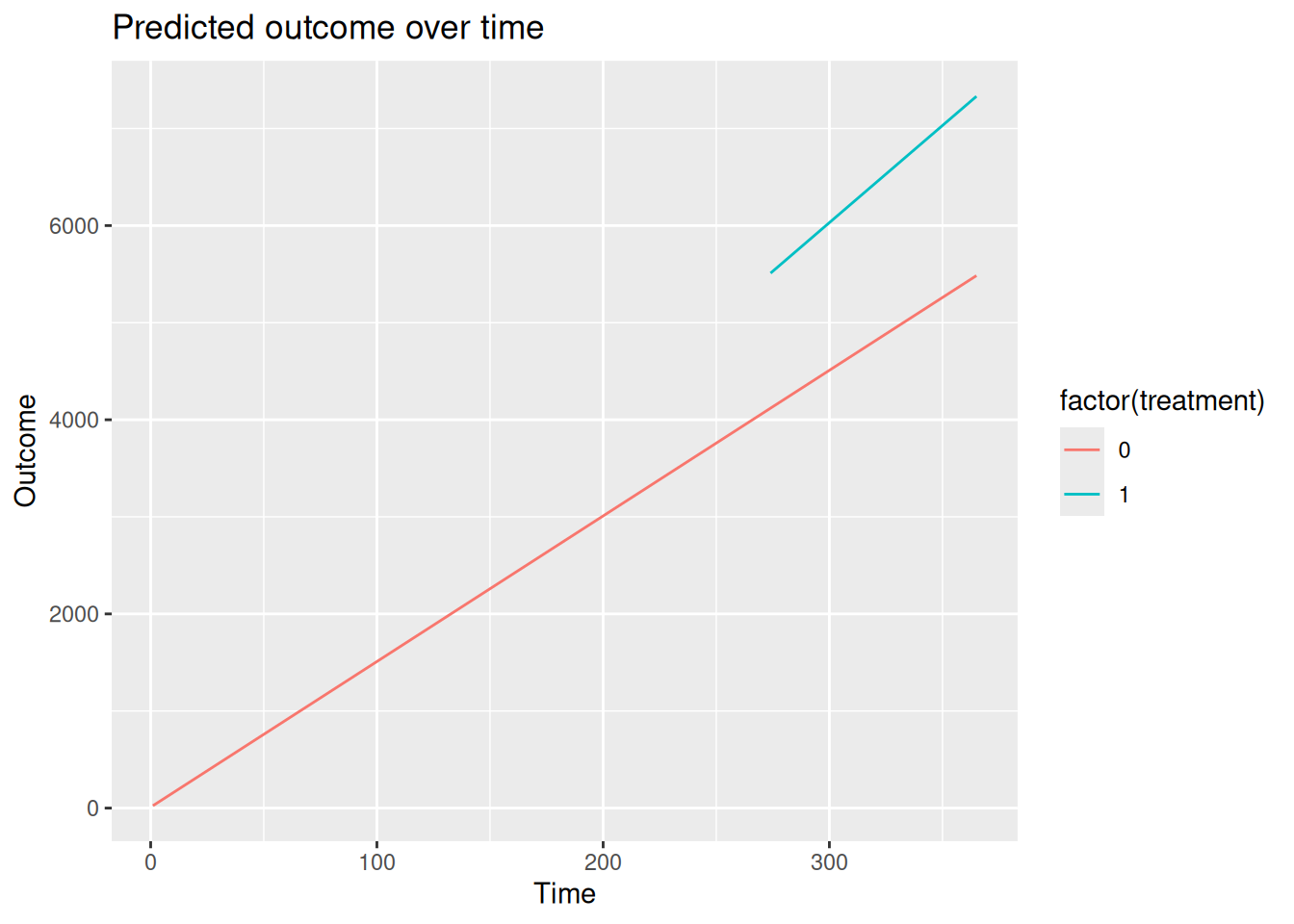
In fact, analysts should not construct this special variable. The problem is that the
timeandtime_sincevariables are related, but when we hard-code them,Rdoes not know the mathematical relationship between them.↩︎